 LABORATORY FOR BIOLOGICAL
LABORATORY FOR BIOLOGICAL
MASS SPECTROMETRY
Home>>Research>>Ion Mobility>>Applications
Ion Mobility Spectrometry
|
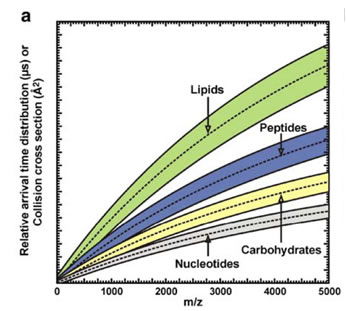
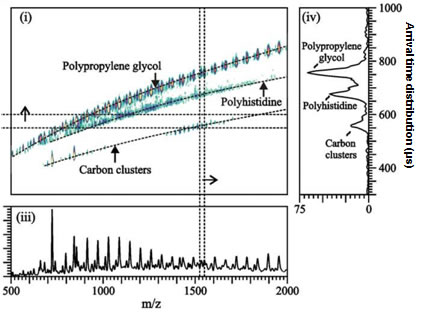
The coupling of ion mobility to mass spectrometry (MS) provides two-dimensional strategy for ultra-fast (us-ms time scale) separation as well as identification of molecular classes based on trend line analysis4. For example, plots of size (collision cross-section) vs mass-to-charge can be related to gas-phase packing efficiency of the ions, thus different chemical classes exhibit different trend lines in the 2D plot. Our group and others successfully utilized IM-MS to screen chemically modified DNA molecules, to distinguish between phosphorylated and nonphosphorylated peptides, to separate mixtures containing lipids, peptides and nucleotides, to characterize synthetic polymers, as well as molecular structure analysis of crude petroleum extracts.
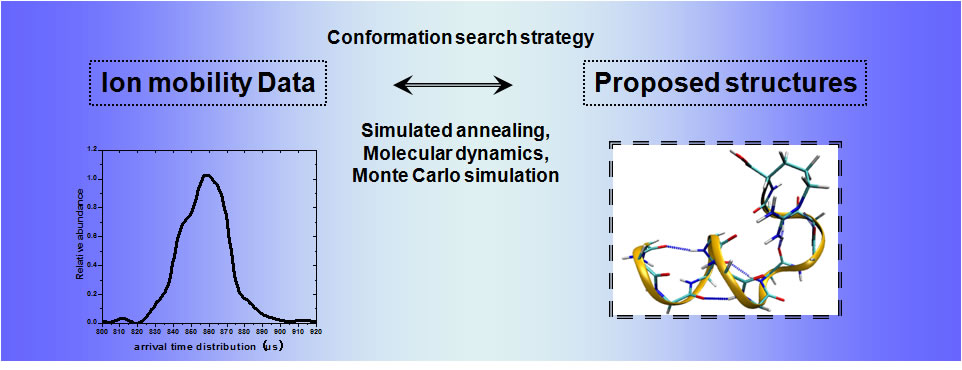
IM-MS is not only used as a separation tool for complex biological mixture, but also used to identify structural isomers and to investigate conformation of gas-phase biological molecules. As a biophysical tool, IM-MS experiments provide empirical collision cross-sections which can be correlated to candidate structures derived from conformation search strategies, i.e. simulated annealing, molecular dynamics.
|
 |