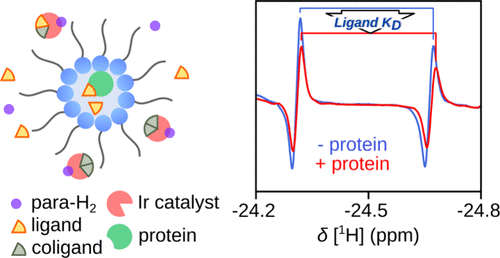
105. Pham, P., Biswas, O. and Hilty, C. Parahydrogen Polarization in Reverse Micelles and Application to Sensing of Protein-Ligand Binding J. Am. Chem. Soc. doi:10.1021/jacs.4c13177. (article, web: December 9, 2024)
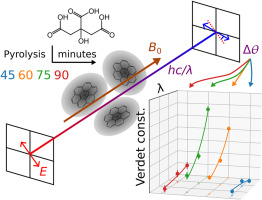
104. Zhang, Z., Savukov, I.* and Hilty, C.* Large Faraday rotation in pyrolysis synthesized carbon dots. Carbon 233 5063–5066 (2024). (article, web: December 4, 2024)
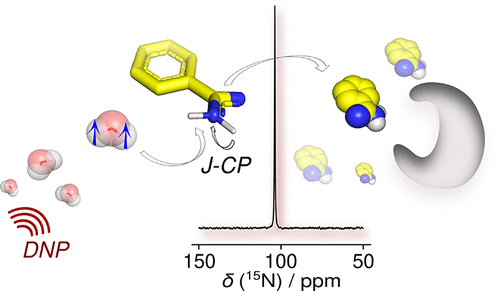
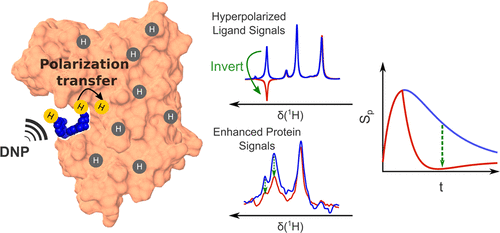
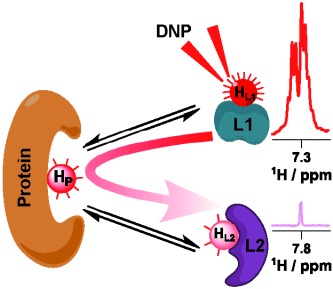
103. Pradhan, N. and Hilty, C. Cross-Polarization of Insensitive Nuclei from Water Protons for Detection of Protein–Ligand Binding. J. Am. Chem. Soc. 146(36) 24754–24758 (2024). (article, web: September 3, 2024)
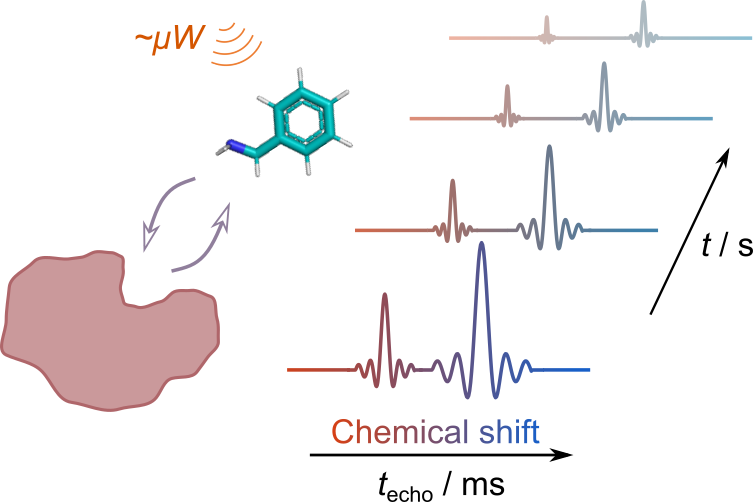
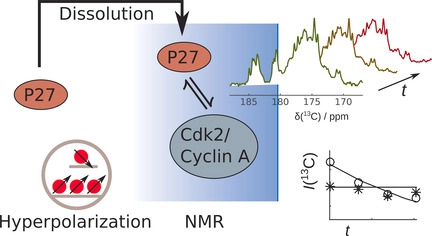
102. Qi, C., Mankinen, O.*, Telkki, V.V. and Hilty, C.* Measuring Protein–Ligand Binding by Hyperpolarized Ultrafast NMR. J. Am. Chem. Soc. 146(8) 5063–5066 (2024). (article, web: February 19, 2024)
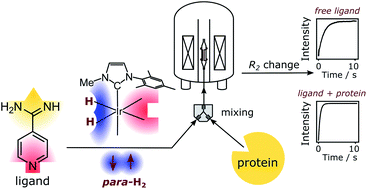
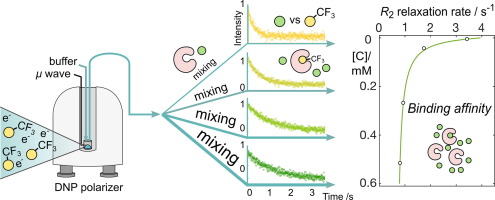
101. Pham, P. and Hilty, C. R2 Relaxometry of SABRE Hyperpolarized Substrates at Low Magnetic Field. Anal. Chem. 95(46) 16911–16917 (2023). (article, web: November 6, 2023)
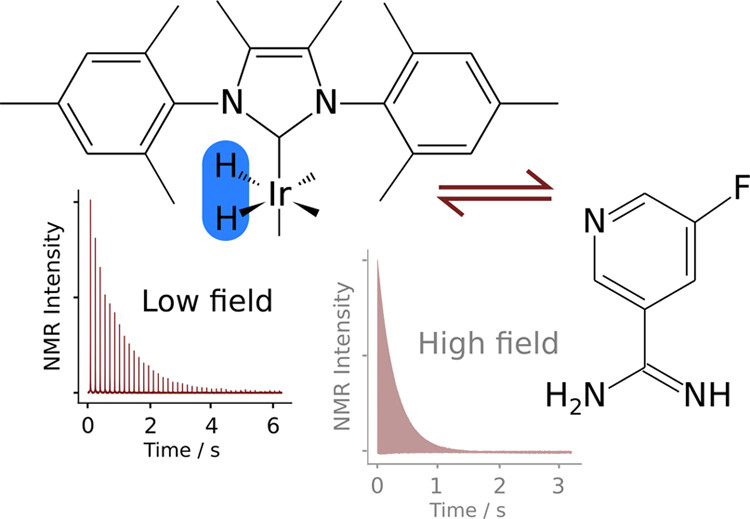
100. Zhang, Z., Gautam, A., Lim, S.M. and Hilty, C. Analysis of Large Data Sets in a Physical Chemistry Laboratory NMR Experiment using Python. J. Chem. Ed. 100(10), 4109–4113 (2023). (article, web: September 19, 2023)
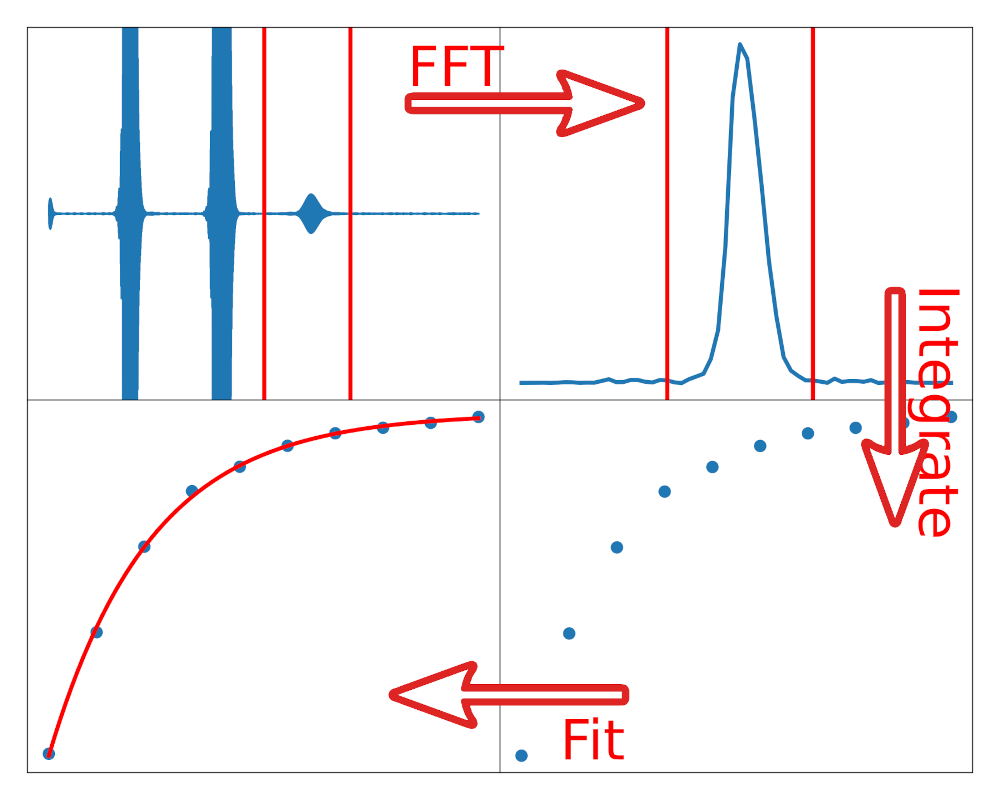
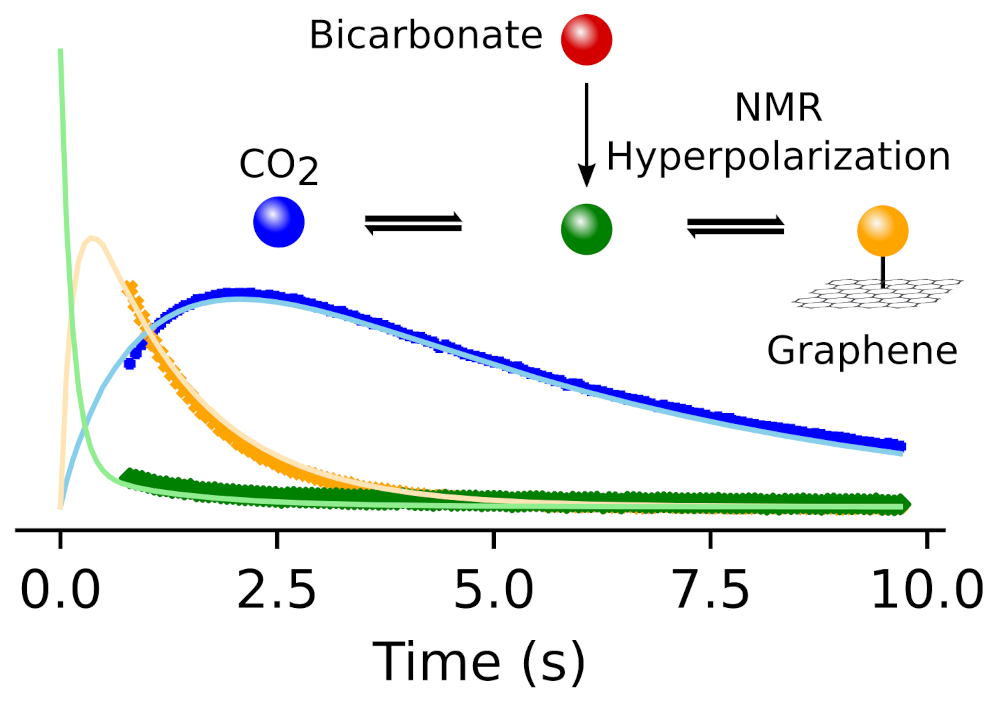
99. Zhang, Z. and Hilty, C. Substrate Interaction of Graphene Quantum Dots in Solution Studied by Hyperpolarized NMR Carbon 215, 118446 (2023). (article, web: September 10, 2023)
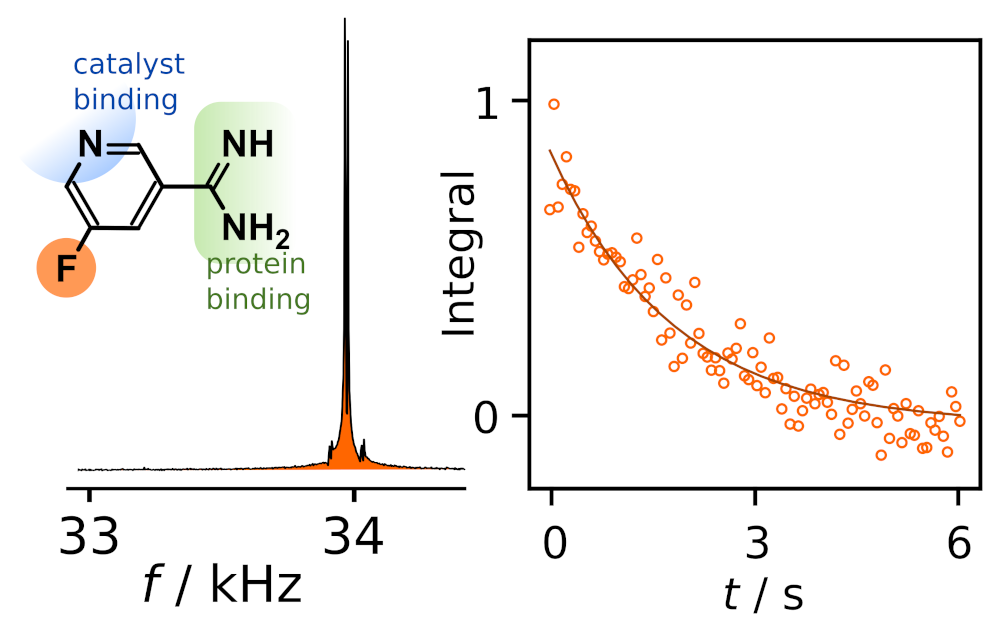
98. Pham, P. and Hilty, C. Biomolecular interactions studied by low-field NMR using SABRE hyperpolarization. Chem. Sci. 14, 10258-10263 (2023). (article, web: September 1, 2023)
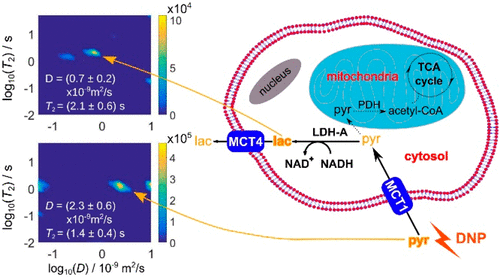
97. Mandal, R., Pham, P. and Hilty, C. Screening of Protein-Ligand Binding using a SABRE Hyperpolarized Reporter. Anal. Chem. 94, 32, 11375–11381 (2022). (article, web: August 3, 2022)
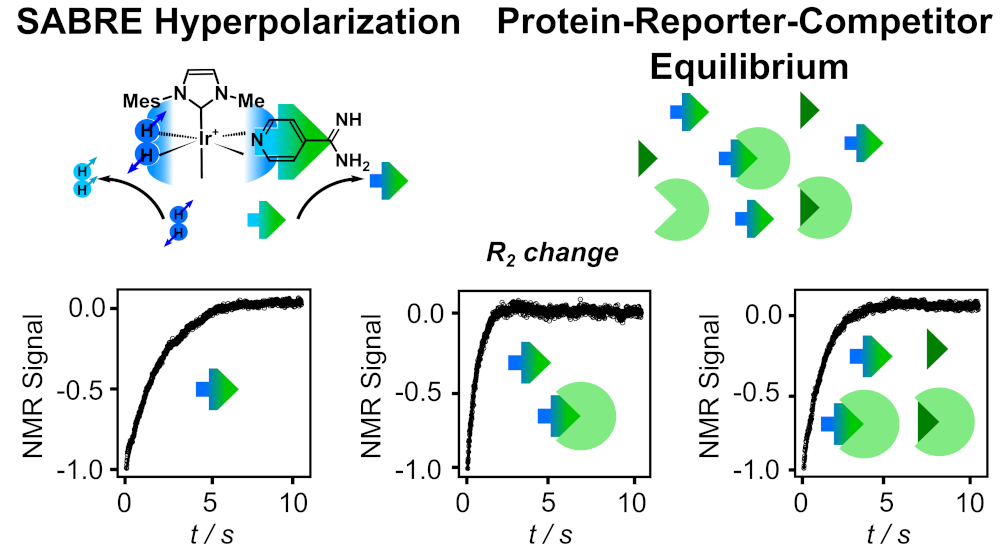
96. Hilty, C.*, Kurzbach, D.* and Frydman, L.* Hyperpolarized water as universal sensitivity booster in biomolecular NMR. Nat. Protoc. doi: https://doi.org/10.1038/s41596-022-00693-8 (2022). (article, web: May, 11 2022)
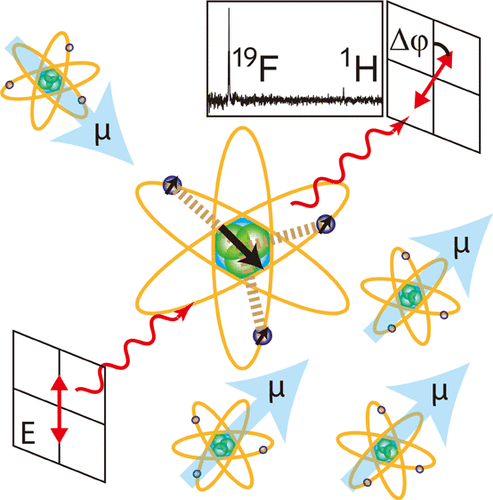
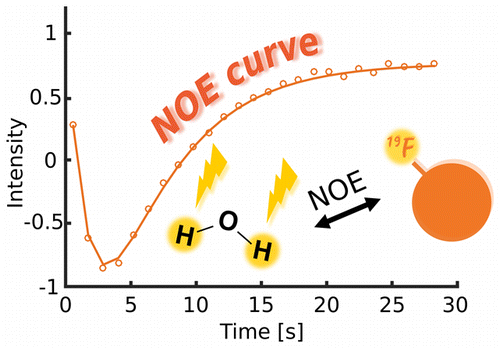
95. Hu, J., Kim, J. and Hilty, C. Detection of Protein-Ligand Interactions by 19F Nuclear Magnetic Resonance Using Hyperpolarized Water. J. Phys. Chem. Lett. 13:3819-3823 (2022). (article, web: April, 24 2022)
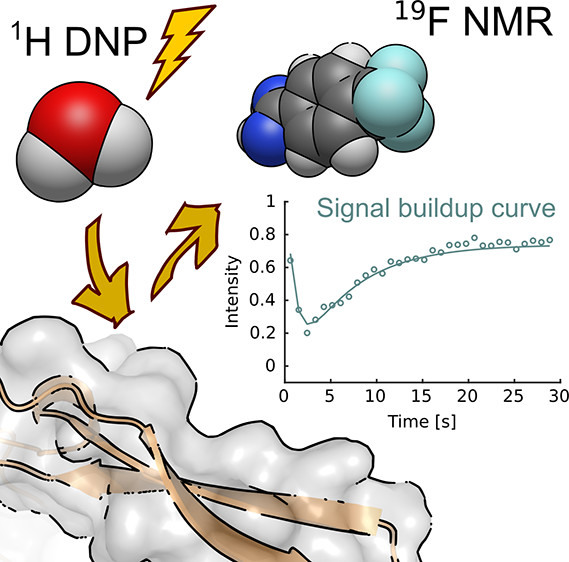
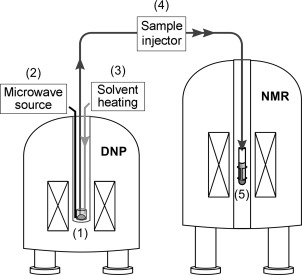
94. Pham, P., Mandal, R., Qi., C. and Hilty, C. Interfacing Liquid State Hyperpolarization Methods with NMR Instrumentation. J. Magn. Reson. Open doi: https://doi.org/10.1016/j.jmro.2022.100052 (2022). (article, web: March 10, 2022)

93. Jaroszewicz, M.J., Liu, M., Kim, J., Zhang, G., Kim, Y., Hilty, C.* and Frydman, L.* Time- and site-resolved kinetic NMR for real-time monitoring of off-equilibrium reactions by 2D spectrotemporal correlations Nat. Commun. 13:833 (2022). (article, web: Feb 11, 2022)
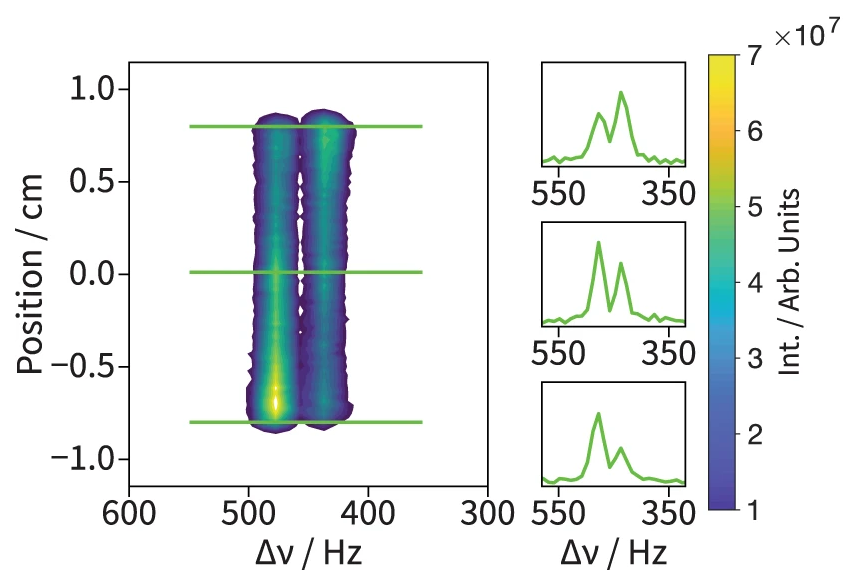
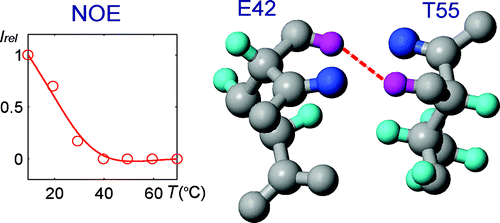
92. Qi, C., Wang, Y. and Hilty, C. Application of Relaxation Dispersion of Hyperpolarized 13C Spins to Protein-Ligand Binding. Angew. Chem. Int. Ed. 60(45): 24018-24021 (2021). (article, web: Sep. 1, 2021)
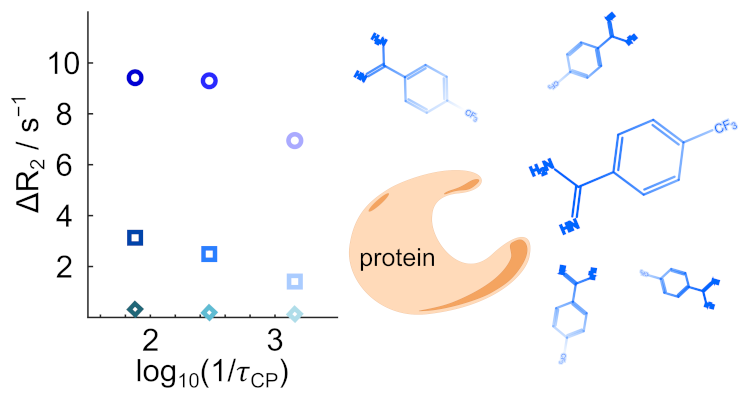
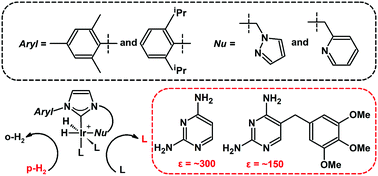
91. Mandal, R., Pham, P. and Hilty, C.
Characterization of protein–ligand interactions
by SABRE. Chem. Sci. 12(39):
12950-12958 (2021).
(article, web: Aug. 31, 2021).
This article is part of the 2021 Chemical
Science HOT Article Collection
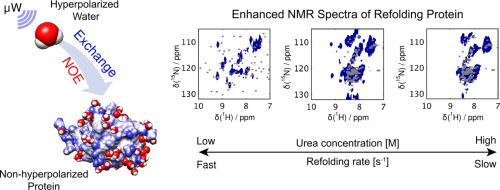
90. Kim, J., Mandal, R. and Hilty, C. 2D NMR Spectroscopy of Refolding RNase Sa using Polarization Transfer from Hyperpolarized Water. J. Magn. Reson. 326:106942 (2021). (article, web: Feb. 18, 2021)
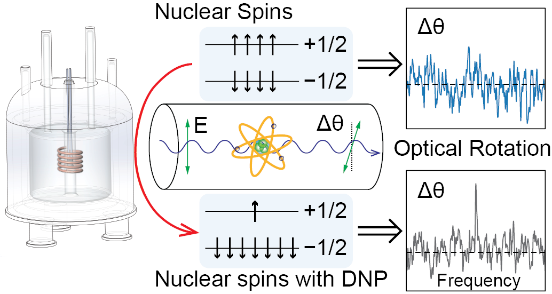
89. Zhu, Y., Hilty, C.* and Savukov,
I.* Dynamic Nuclear Polarization Enhanced Nuclear
Spin Optical Rotation. Angew. Chem. Int.
Ed. 60(16): 8823-8826 (2021).
(article, web: Jan. 18, 2021)
This article has been designated a "Hot Paper"
by the journal.
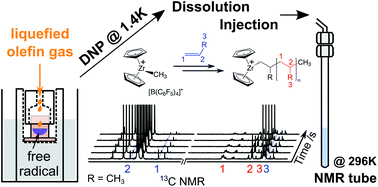
88. Kim, Y., Samouei, H. and Hilty, C.
Polyolefin Catalysis of Propene, 1-Butene and
Isobutene Monitored Using Hyperpolarized NMR.
Chem. Sci. 12(8): 2823-2828 (2021).
(article, web: Jan. 4, 2021)
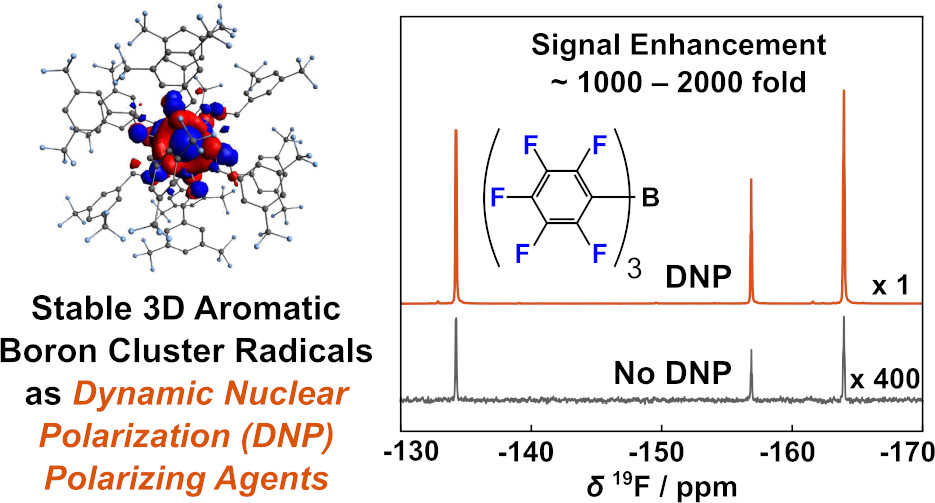
87. Kim, Y., Kubena, R., Axtell, J.,
Samouei, H., Pham, P., Stauber, J., Spokoyny, A.*
and Hilty, C.* Dynamic Nuclear Polarization using
3D Aromatic Boron Cluster Radicals. J. Phys.
Chem. Lett. 12(1): 13-18 (2021).
(article, web: Dec. 9, 2020)
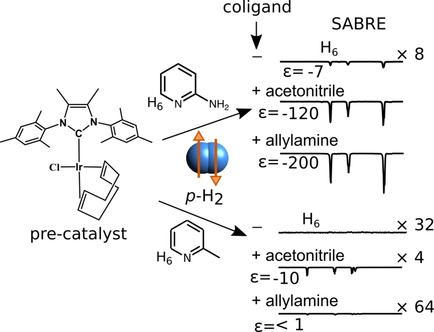
86. Pham, P. and Hilty, C. Tunable
iridium catalyst designs with bidentate
N-heterocyclic carbene ligands for SABRE
hyperpolarization of sterically hindered
substrates. Chem. Commun. 56(98):
15466-15469 (2020).
(article, web: Nov. 18, 2020)
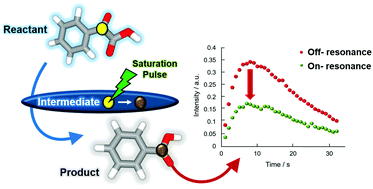
85. Kim, J., Kim, Y., Luu, Q.S.,
Kim, J., Qi, C., Hilty, C.* and Lee, Y.* Indirect
detection of intermediate in decarboxylation
reaction of phenylglyoxylic acid by
hyperpolarized 13C NMR. Chem.
Commun. 56(95): 15000-15003 (2020).
(article, web: Nov. 8, 2020)
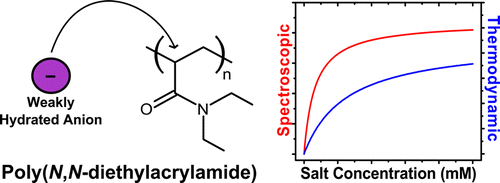
84. Wang, Y. and Hilty, C.
Amplification of Nuclear Overhauser Effect by
Hyperpolarization for Screening of Ligand Binding
to Immobilized Target Proteins. Anal.
Chem. 92(20): 13718-13723 (2020).
(article, web: Sep. 8, 2020)

83. Mandal, R., Pham, P. and Hilty, C.
Nuclear Spin Hyperpolarization of NH2
and CH3-Substituted Pyridine and
Pyrimidine Moieties by SABRE. ChemPhysChem
21(19): 2166-2172 (2020). (article,
web: Aug. 12, 2020)
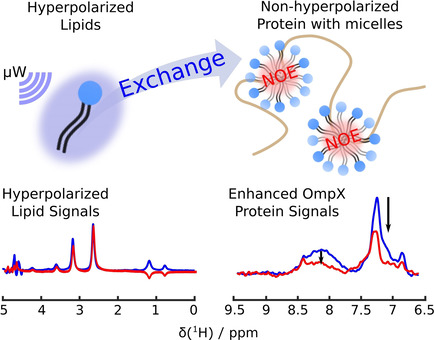
82. Kim, J., Mandal, R. and
Hilty, C. Characterization of Membrane
Protein-Lipid Interactions in Unfolded OmpX with
Enhanced Time Resolution by Hyperpolarized NMR.
ChemBioChem 21(19): 2861-2867
(2020).
(article, web: May 17, 2020)
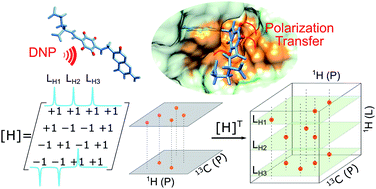
81. Wang, Y., Kim, J. and Hilty, C.
Determination of protein-ligand binding modes
using fast multi-dimensional NMR with
hyperpolarization. Chem. Sci.
11(23):5935-5943 (2020).
(article, web: May 6, 2020).
This article is part of the 2020 Chemical
Science HOT Article Collection
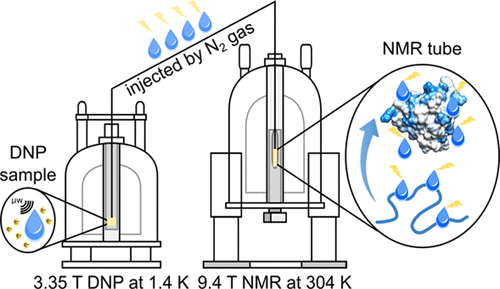
80. Kim, J., Mandal, R. and Hilty, C.
Observation of Fast Two-Dimensional NMR Spectra
during Protein Folding Using Polarization
Transfer from Hyperpolarized Water. J. Phys.
Chem. Lett. 10(18): 5463-5467 (2019).
(article, web: Aug 23, 2019)
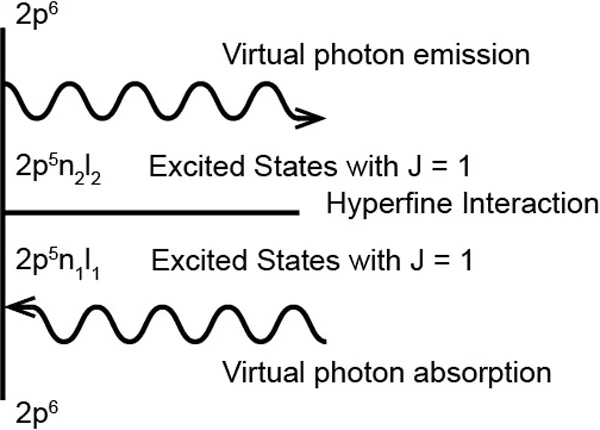
79. Savukov, I., Filin, D., Zhu, Y.,
Castro, R. and Hilty, C. Calculations of neon
nuclear-spin optical rotation, Verdet and
hyperfine constants with
configuration-interaction many-body perturbation
theory. Eur. Phys. J. D 73(156):
2419-2427 (2019).
(article, web: Jul 25, 2019)
78. Wang, Y. and Hilty, C.
Determination of Ligand Binding Epitope
Structures Using Polarization Transfer from
Hyperpolarized Ligands. J. Med. Chem.
62(5): 2419-2427 (2019).
(article, web: Feb 4, 2019)
77. Kim, Y. and Hilty, C. Applications
of Dissolution-DNP for NMR Screening. In:
Wand, J. (ed) Methods in Enzymology, Biological
NMR Part B, Volume 615, 501-526 (2019). ISSN
0076-6879.
(article, web: Dec. 4, 2018)
76. Zhang, G., Ahola, S., Lerche, M.,
Telkki, V.V. and Hilty, C. Identification of
Intra- and Extracellular Metabolites in Cancer
Cells Using 13C Hyperpolarized
Ultrafast Laplace NMR. Anal. Chem.
90(18): 11131-11137 (2018).
(article, web: Aug 20, 2018)
75. Kim, Y.,Liu, M., and Hilty,
C. Determination of binding affinities using
hyperpolarized NMR with simultaneous 4-channel
detection. J. Magn. Reson. 295:
80-86 (2018).
(article, web: Aug 13, 2018)
74. King, J.N., Fallorina, A., Yu, J.,
Zhang, G., Telkki, V.V., Hilty, C. and Meldrum,
T. Probing molecular dynamics with hyperpolarized
ultrafast Laplace NMR using a low-field,
single-sided magnet. Chem. Sci. 9:
6143-6149 (2018).
(article, web: June 28, 2018)
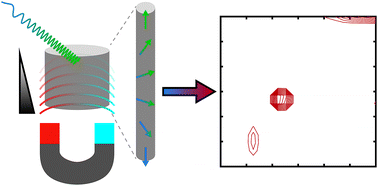
73. Zhu, Y., Gao, Y., Rodocker, S.,
Savukov, I. and Hilty, C. Multinuclear Detection
of Nuclear Spin Optical Rotation at Low Field.
J. Phys. Chem. Lett. 9(12):
3323-3327 (2018).
(article, web: May 22, 2018)
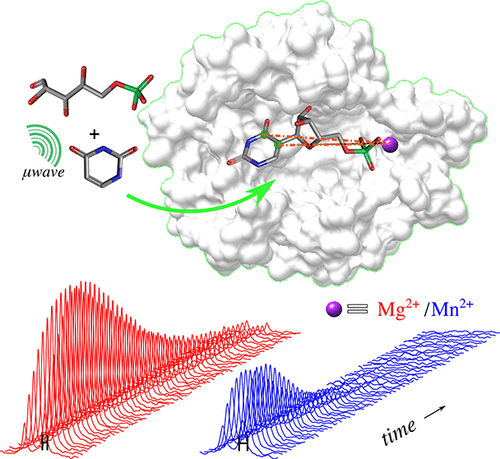
72. Liu, M., Zhang, G., Mahanta, N.,
Lee, Y. and Hilty, C. Measurement of Kinetics and
Active Site Distances in Metalloenzyme
Using Paramagnetic NMR with 13C
Hyperpolarization. J. Phys. Chem. Lett.
9(9): 2218-2221 (2018).
(article, web: April 6, 2018)
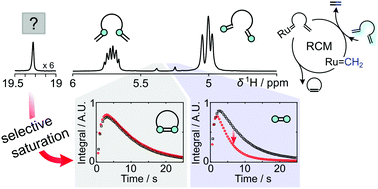
71. Kim, Y., Chen, C.H. and Hilty, C.
Direct observation of Ru-alkylidene forming into
ethylene in ring-closing metathesis from
hyperpolarized 1H NMR. Chem.
Commun. 54: 4333-4336 (2018).
(article, web: April 6, 2018)
70. Zhang, G. and Hilty, C.
Applications of Dissolution Dynamic Nuclear
Polarization in Chemistry and Biochemistry.
Magn. Reson. Chem. 56(7): 566-582
(2018).
(article, web: March 30, 2018)
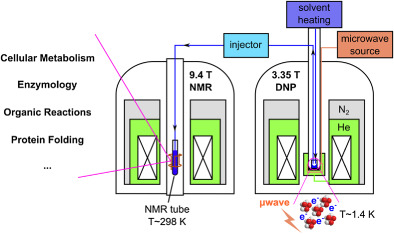
69. Chen, C.H., Wang Y. and Hilty, C. Intermolecular Interactions Determined by NOE Build-up in Macromolecules from Hyperpolarized Small Molecules. Methods 138-139: 69-75 (2018). (article, web: February 19, 2018)
68. Liu, M. and Hilty, C. Metabolic
Measurements of Nonpermeating Compounds in Live
Cells Using Hyperpolarized NMR. Anal.
Chem. 90(2): 1217-1222 (2018).
(article, web: Dec 11, 2017)
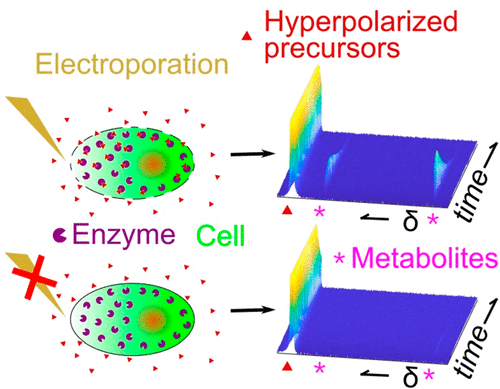
67. Liu, M., Kim, Y. and Hilty, C.
Characterization of Chemical Exchange Using
Relaxation Dispersion of Hyperpolarized Nuclear
Spins. Anal. Chem. 89(17):
9154-9158 (2017).
(article, web: Jul 17, 2017)
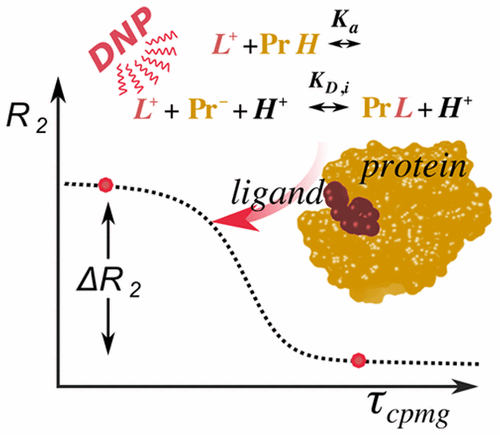
66. Kim, J., Liu, M. and Hilty, C.
Modeling of Polarization Transfer Kinetics in
Protein Hydration Using Hyperpolarized Water.
J. Phys. Chem. B 121(27): 6492-6498
(2017).
(article, web: Jun 14, 2017)
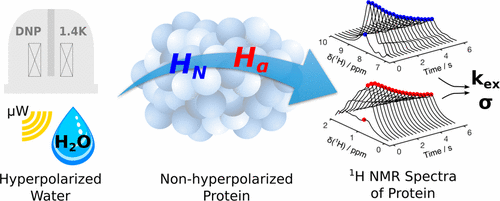
65. Ragavan, M., Iconaru, L.I., Park,
C.G., Kriwacki, R.W. and Hilty, C. Real-Time
Analysis of Folding upon Binding of a Disordered
Protein by using Dissolution DNP NMR
Spectroscopy. Angew. Chem. Int. Ed.
56(25): 7070-7073 (2017).
(article, web: May 16, 2017)
64. Kim, Y., Liu, M. and Hilty, C.
Parallelized Ligand Screening Using Dissolution
Dynamic Nuclear Polarization. Anal. Chem.
88(22): 11178-11183 (2016).
(article, web: Oct 10, 2016)

63. Zhang, G., Schilling, F.,
Glaser, S.J. and Hilty, C. Reaction Monitoring
Using Hyperpolarized NMR with Scaling of
Heteronuclear Couplings by Optimal Tracking.
J. Magn. Reson. 272: 123-128
(2016).
(article, web: Sep 14, 2016)
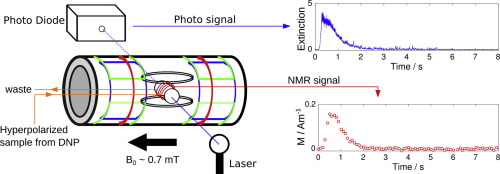
62. Zhu, Y., Chen, C.H., Wilson, Z.,
Savukov, I., and Hilty, C. Milli-tesla NMR and
spectrophotometry of liquids hyperpolarized by
dissolution dynamic nuclear polarization. J.
Magn. Reson. 270: 71-76 (2016).
(article, web: July 13, 2016)
61. Wang, Y., Ragavan, M., and Hilty, C. Site specific polarization transfer from a hyperpolarized ligand of dihydrofolate reductase. J. Biomol. NMR: 41-48 (2016). (article, web: May 17, 2016)
60. Kim, J., Liu, M., Chen, H.Y. and
Hilty, C. Determination of Intermolecular
Interactions using Polarization Compensated
Heteronuclear Overhauser Effect of Hyperpolarized
Spins. Anal. Chem. 87(21):
11092-10987 (2015).
(article, web: Oct 1, 2015)
59. Ahola, S., Zhivonitko, V.V., Mankinen, O., Zhang, G., Kantola, A.M., Chen, H.Y., Hilty, C., Koptyug, I.V. and Telkki, V.V. Ultrafast multidimensional Laplace NMR for a rapid and sensitive chemical analysis. Nat. Commun. 6: 8363 (2015). (article, web: Sep 18, 2015)
58. Min, H., Sekar, G. and Hilty, C.
Polarization Transfer from Ligands Hyperpolarized
by Dissolution Dynamic Nuclear Polarization for
Screening in Drug Discovery. ChemMedChem
10(9): 1559-1563 (2015).
(article, web: Aug 4, 2015)
57. Ardenkjaer-Larsen, J.H., Boebinger,
G.S., Comment, A., Duckett, S., Edison, A.S.,
Engelke, F., Griesinger, C., Griffin, R.G.,
Hilty, C., Maeda, H., Parigi, G., Prisner, T.,
Ravera, E., Bentum, J.V., Vega, S., Webb, A.,
Luchinat, C., Schwalbe, H. and Frydman, L. Facing
and Overcoming Sensitivity Challenges in
Biomolecular NMR Spectroscopy Angew. Chem.
Int. Ed. 54(32): 9162-9185 (2015).
(Review article).
(article, web: Jul 1, 2015)
56. Chen, H.Y. and Hilty, C.
Implementation and Characterization of Flow
Injection in Dissolution Dynamic Nuclear
Polarization NMR Spectroscopy.
ChemPhysChem 16(12): 2646-2652
(2015).
(article, web: Jul 2, 2015)
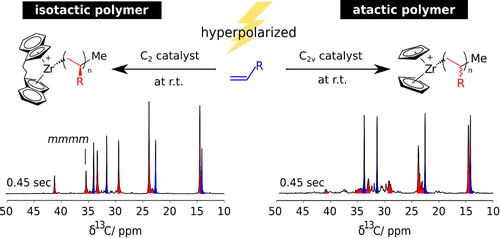
55. Chen, C.H., Shih, W.C. and Hilty,
C. In-Situ Determination of Tacticity,
Deactivation and Kinetics in
[rac-(C2H4(1-indenyl)2)ZrMe][B(C6F5)4]
and
[(Cp)2ZrMe][B(C6F5)4]-Catalyzed
Polymerization of 1-Hexene using 13C
Hyperpolarized NMR. J. Am. Chem. Soc.
137(21): 6965-6971(2015).
(article, web: May 11, 2015)
54. Rembert, K.B., Okur, H.I., Hilty,
C. and Cremer, P. An NH Moiety Is Not Required
for Anion Binding to Amides in Aqueous Solution.
Langmuir 31(11): 3459-3464 (2015).
(article, web: Mar 12, 2015)
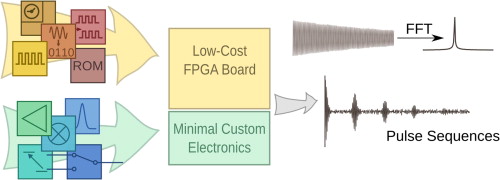
53. Chen, H.Y., Kim, Y., Nath, P., and
Hilty, C. An Ultra-Low Cost NMR Device with
Arbitrary Pulse Programming. J. Magn.
Reson. 255: 100-105 (2015).
(article, web: Mar 10, 2015)
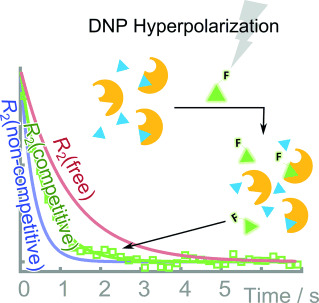
52. Kim, Y. and Hilty, C. Affinity
Screening Using Competitive Binding with
Fluorine-19 Hyperpolarized Ligands. Angew.
Chem. Int. Ed. 54(16): 4941-4944
(2015).
(article, Feb 20, 2015)
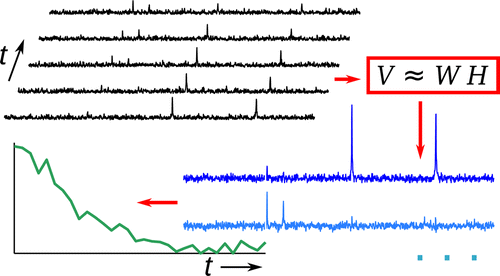
51. Hilty, C. and Ragavan, M.
Application of Blind Source Separation to
Real-Time Dissolution Dynamic Nuclear
Polarization. Anal. Chem. 87(2):
1004-1008 (2015). (article,
web: Dec 15, 2014)
50. Hladilkova, J., Hyeda, J., Rembert,
K., Okur, H., Kurra, Y., Liu, W.R., Hilty, C.,
Cremer, P., and Jungwirth, P. Effects of
End-Group Termination on Salting-Out Constants
for Triglycine. J. Phys. Chem. Lett.
4: 4096-4073 (2013). (article,
web: Nov 19, 2013)

49. Chen, H.Y., Ragavan, M. and
Hilty, C. Protein Folding Studied by Dissolution
Dynamic Nuclear Polarization. Angew. Chem.
Int. Ed. 52(35): 9192-9195 (2013).
(article, web: Jul 15, 2013)
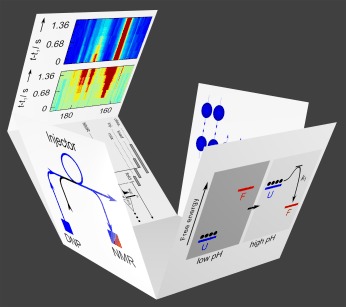
48. Chen, H.Y., Hilty, C.
Hyperpolarized Hadamard Spectroscopy Using Flow
NMR. Anal. Chem. 85(15): 7385-7390
(2013). (article,
web: Jul 18, 2013)
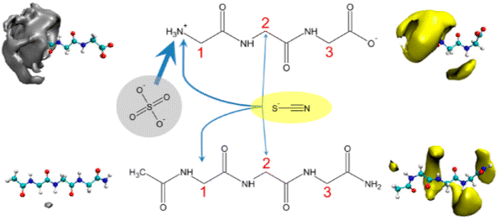
47. Paterova, J., Rembert, K., Heyda,
J., Kurra, Y., Okur, H., Liu, W.R., Hilty, C.,
Cremer, P., and Jungwirth, P. Reversal of the
Hofmeister Series: Specific Ion Effects on
Peptides. J. Phys. Chem. B 117(27):
8150-8158 (2013). (article,
web: Jun 14, 2013)
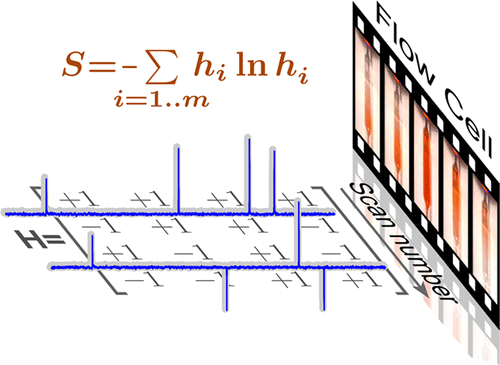
46. Savukov, I.M., Chen, H.Y.,
Karaulanov, T. and Hilty, C. Method for accurate
measurements of nuclear-spin optical rotation for
applications in correlated optical-NMR
spectroscopy. J. Magn. Reson. 232:
31-38 (2013). (article,
web: Apr 24, 2013)

45. Lee, Y., Heo, G.S., Zeng, H.,
Wooley, K. and Hilty, C. Detection of Living
Anionic Species in Polymerization Reaction using
Hyperpolarized NMR. J. Am. Chem. Soc.
135(12): 4636-4639 (2013). (article,
web: Mar 13, 2013)
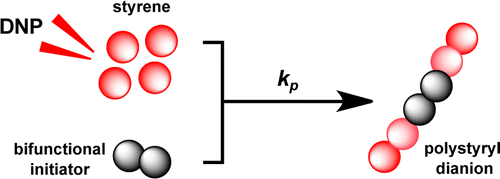
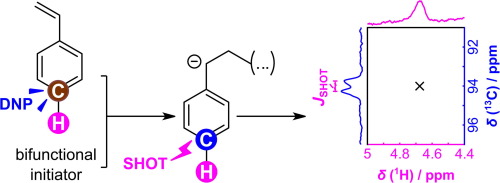
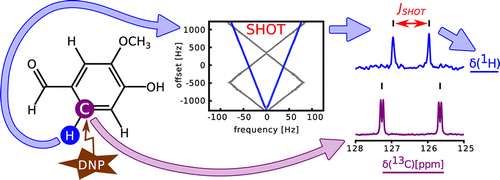
44. Zhang, G., Schilling, F.,
Glaser, S. J. and Hilty, C. Chemical Shift
Correlations from Hyperpolarized NMR using a
single SHOT. Anal. Chem. 85(5):
2875-2881 (2013). (article,
web: Feb 14, 2013)
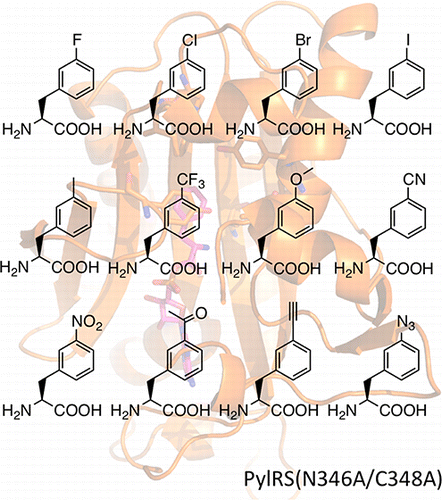
43. Wang, Y.S., Fang, X., Chen, H.Y.,
Wu, B., Wang, Z. W., Hilty, C. and Liu, W. R.
Genetic Incorporation of Twelve
meta-Substituted Phenylalanine Derivatives
Using a Single Pyrrolysyl-tRNA Synthetase Mutant.
ACS Chem. Biol. 8(2): 405-415
(2013). (article,
web: Nov 19, 2012)
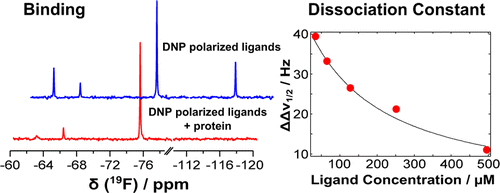
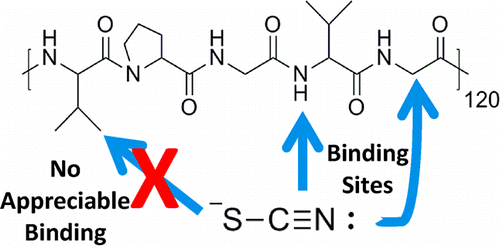
42. Lee, Y., Zeng, H.,
Ruedisser, S., Gossert, A.D. and Hilty, C.
Nuclear Magnetic Resonance of Hyperpolarized
Fluorine for Characterization of Protein-Ligand
Interactions. J. Am. Chem. Soc.
134(42): 17448-17451 (2012). (article,
web: Oct 9, 2012). Highlighted in
Nature SciBX.
41. Rembert, K.B., Paterová, J., Heyda,
J., Hilty, C., Jungwirth, P. and Cremer, P.S.
Molecular Mechanisms of Ion-Specific Effects on
Proteins. J. Am. Chem. Soc.
134(24): 10039-10046 (2012). (article,
web: Jun 11, 2012)
See also coverage in
C&E News.
40. Lee, Y., Zeng, H. Mazur, A.,
Wegstroth, M., Carlomagno, T., Reese, M., Lee,
D., Becker, S., Griesinger, C. and Hilty, C.
Hyperpolarized Binding Pocket Nuclear Overhauser
Effect for Determination of Competitive Ligand
Binding. Angew. Chem. Int. Ed.
51(21): 5179-5182 (2012).
(article, web: Apr 12, 2012)
39. Hwang, S., and Hilty, C. Folding of
a tryptophan zipper peptide investigated based on
Nuclear Overhauser effect and thermal
denaturation. J. Phys. Chem. B
115(51): 15355-15361 (2011). (article,
web: Dec. 6, 2011)
38. Ragavan, M., Chen, H.Y., Sekar, G.
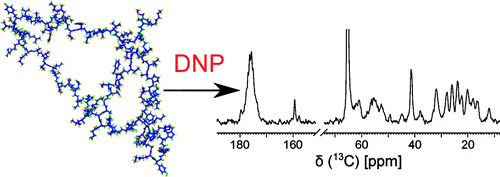
and Hilty, C. Solution NMR of Polypeptides
Hyperpolarized by Dynamic Nuclear Polarization.
Anal. Chem. 83(15): 6054-6059
(2011). (article,
web: Jun 30, 2011)
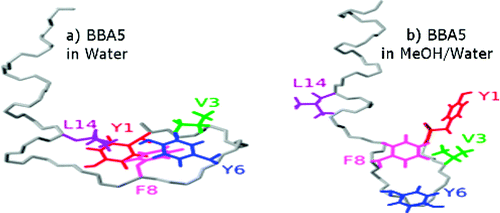
37. Hwang, S., Shao, Q.,
Williams, H., Hilty, C. and Gao, Y.Q. Methanol
Strengthens Hydrogen Bonds and Weakens
Hydrophobic Interactions in Proteins - A Combined
Molecular Dynamics and NMR study. J. Phys.
Chem. B 115(20): 6653-6660 (2011).
(article,
web: May 2, 2011)
36. Bowen, S., Sekar, G. and Hilty, C. Rapid Determination of Biosynthetic Pathways using Fractional Isotope Enrichment and High-Resolution Dynamic Nuclear Polarization Enhanced NMR. NMR Biomed. 24(4): 1016-1022 (2011). (article, web: Mar 8, 2011)
35. Hwang, S. and Hilty, C. Folding determinants of disulfide bond forming protein B explored by solution NMR Spectroscopy. Proteins 79(5):1365-1375 (2011). (article, web: Feb 18, 2011)
34. Chen, H.Y., Lee, Y., Bowen, S. and
Hilty, C. Spontaneous emission of NMR signals in
hyperpolarized proton spin systems. J. Magn.
Reson. 208(2): 204-209 (2011).
(article, web: Nov 11, 2010)
33. Choutko, A., Glättli, A., Fernández, C., Hilty, C., Wüthrich, K. and van Gunsteren, W.F. Membrane protein dynamics in different environments: Simulation study of the outer membrane protein X (OmpX) in a lipid bilayer and in a micelle. Eur. Biophys. J. 40(1): 39-58 (2011). (article, web: Oct 5, 2010)
32. Zeng, H., Lee, Y. and Hilty, C.
Quantitative rate determination by DNP enhanced
NMR of a Diels-Alder reaction. Anal. Chem.
82(21): 8897-8902 (2010). (article,
web: Oct 13, 2010)
31. Hilty, C. and Bowen, S.
Applications of dynamic nuclear polarization to
the study of reactions and reagents in organic
and biomolecular chemistry. Org. Biomol.
Chem. 8(15): 3361-3365 (2010).
(Emerging area article).
(article, web: Jun 7, 2010)
30. Bowen, S., Hilty, C. Rapid sample
injection for hyperpolarized NMR spectroscopy
Phys. Chem. Chem. Phys. 12(22):
5766-5770 (2010).
(article, web: May 4, 2010)
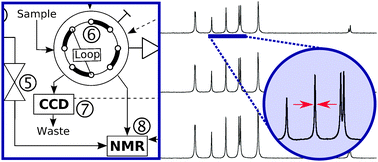
29. Hilty, C., Bowen, S. A NMR
experiment based on off-the-shelf digital data
acquisition equipment. J. Chem. Edu.
87(7): 747-749 (2010). (article,
web: May 19, 2010)
28. Zeng, H., Bowen, S., Hilty, C. Sequentially acquired two-dimensional NMR spectra from hyperpolarized sample. J. Magn. Reson. 199 (2): 159-165 (2009). (article, web: Apr 24, 2009)
27. Bowen, S., Hilty, C. Temporal chemical shift correlations in reactions studied by hyperpolarized NMR. Anal. Chem. 81 (11): 4543-4547 (2009). (article, web: Apr 23, 2009)
26. Lee, D., Walter, K. F. A.,
Brückner, A.K., Hilty, C., Becker, S.,
Griesinger, C. Bilayer in Small Bicelles Revealed
by Lipid-Protein Interactions Using NMR
Spectroscopy. J. Am. Chem. Soc.
130(42): 13822-13823 (2008). (article,
web: Sep 26, 2008)

25. Bowen, S., Zeng, H., Hilty, C. Chemical Shift Correlations from Hyperpolarized NMR by Off-Resonance Decoupling. Anal. Chem. 80(15): 5794-5798 (2008). (article, web: Jul 8, 2008)
24. Bowen, S., Hilty, C. Time-resolved
dynamic nuclear polarization enhanced NMR
Spectroscopy. Angew. Chem. Int. Ed.
47(28): 5235-5237 (2008).
(article, web: Jun 2, 2008)
This article has been designated a "Hot Paper"
by the journal. See also coverage in
C&E News.
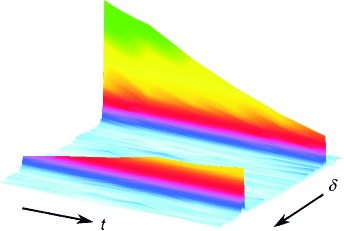
23. Verpillat, F., Ledbetter, M.P., Xu, S., Michalak, D.J., Hilty, C., Bouchard, L.-S., Antonijevic, S., Budker, D., Pines, A. Remote detection of nuclear magnetic resonance with an anisotropic magnetoresistive sensor. P. Natl. Acad. Sci. USA 105(7): 2271-2273 (2008). (article, web: Feb 11, 2008)
22. Telkki, V.V., Hilty, C., Garcia,
S., Harel, E., Pines, A. Quantifying the
Diffusion of a Fluid through Membranes by Double
Phase Encoded Remote Detection Magnetic Resonance
Imaging. J. Phys. Chem. B 111(50):
13929-13936 (2007). (article,
web: Nov 15, 2007)
Cover article
21. Granwehr, J., Harel, E., Hilty, C., Garcia, S., Chavez, L., Pines, A., Sen, P.N., Song, Y.Q. Dispersion measurements using time-of-flight remote detection MRI. Magn. Reson. Imaging 25(4): 449-452 (2007). (article, web: Jan 22, 2007)
20. Koptyug, I.V., Kovtunov, K.V., Burt, S.R., Anwar, M.S., Hilty, C., Han, S.I., Pines, A. and Sagdeev, R.Z. para-Hydrogen-Induced Polarization in Heterogeneous Hydrogenation Reactions. J. Am. Chem. Soc. 129(17): 5580-5586 (2007). (article, web: Apr 5, 2007)
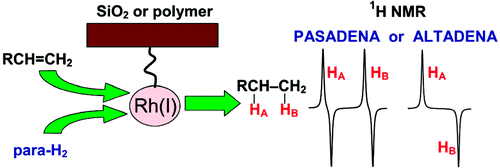
19. Anwar, M.S., Hilty, C., Chu, C., Bouchard, L.S., Pierce, K.L. and Pines, A. Spin coherence transfer in chemical transformations monitored by remote detection NMR. Anal. Chem. 79(7): 2806-2811 (2007). (article, web: Mar 3, 2007)
18. Harel, E., Hilty, C., Koen, K., McDonnell, E.E. and Pines, A. Time-of-Flight Flow Imaging of Two-Phase Flow inside a Microfluidic Chip. Phys. Rev. Lett. 98: 017601 (2007). (article, web: Jan 5, 2007)
17. Schröder, L., Lowery, T.J., Hilty,
C., Wemmer, D.E. and Pines, A. Targeted Molecular
Imaging using Magnetic Resonance of
Hyperpolarized Noble Gas. Science
314: 446-449 (2006). (article,
web: Oct 20, 2006)
See
press release,
Perspectives, and
Faculty of 1000 (Biology)
16. Baker, K.A., Hilty, C., Peti, W., Prince, A., Pfaffinger, P.J., Wider, G., Wüthrich, K. and Choe, S. NMR-derived dynamic aspects of N-type inactivation of a Kv channel suggest a transient interaction with the T1 domain Biochemistry 45(6): 1663-1672 (2006). (article, web: Jan 20, 2006)
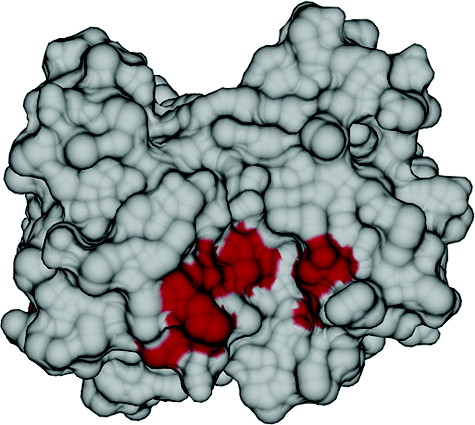
15. Hilty, C., Lowery, T.J., Wemmer,
D.E. and Pines, A. Spectrally Resolved Magnetic
Resonance Imaging of the Xenon Biosensor.
Angew. Chem. Int. Ed. 45(1): 70-73
(2006).
(article, web: Nov 28, 2005)
This article was designated a VIP and
highlighted as "frontispiece".
14. Lee, D., Hilty, C., Wider, G. and Wüthrich, K. Efficient Correlation Time Measurements for Macromolecules from NMR Relaxation Interference. J. Magn. Reson. 178: 72-76 (2006). (article, web: Sep 26, 2005)
13. McDonnell, E., Han, S.I., Hilty, C., Pierce, K.L. and Pines, A. NMR analysis on microfluidic devices by remote detection. Anal. Chem. 77: 8109-8114 (2005). (article, web: Nov 17, 2005)
12. Hilty, C., McDonnell, E., Granwehr,
J., Pierce, K., Han, S.I., and Pines, A.
Microfluidic gas flow profiling using
hyperpolarized xenon and remote detection. P.
Natl. Acad. Sci. USA 102(42):
14960-14963 (2005). (article,
web: Oct 18, 2005)
See
press release and
news article
11. Seebach, D., Mathad, R.I., Kimmerlin, T., Mahajan, Y.R., Bindschadler, P., Rueping, M., Jaun, B., Hilty, C. and Etezady-Esfarjani, T. NMR-Solution Structures in Methanol of an α-Heptapeptide, of a β3/β2-Nonapeptide, and of an all-β3-Icosapeptide Carrying the 20 Proteinogenic Side Chains. Helv. Chim. Acta 88(7): 1969-1982 (2005). (article, web: Jul 20, 2005)
10. Hilty, C., Wider, G., Fernández, C. and Wüthrich, K. Membrane protein-lipid interactions in mixed micelles studied by NMR spectroscopy with the use of paramagnetic reagents. ChemBioChem 5(4): 467-473 (2004). (article, web: Apr 1, 2004)
9. Tafer, H., Hiller, S., Hilty, C., Fernández, C. and Wüthrich, K. Nonrandom Structure in the Urea-Unfolded Escherichia coli Outer Membrane Protein X (OmpX). Biochemistry 43(4): 860-869 (2004). (article, web: Jan 8, 2004)
8. Fernández, C., Hilty, C., Wider, G.,
Güntert, P. and Wüthrich, K. NMR structure of the
integral membrane protein OmpX. J. Mol.
Biol. 336(5): 1211-1221 (2003).
(article, web: Feb 19, 2004)
See
Faculty of 1000 (Biology)
7. Hilty, C., Wider, G., Fernández, C. and Wüthrich, K. Stereospecific assignments of the isopropyl methyl groups of the membrane protein OmpX in DHPC micelles. J. Biomol. NMR 27(4): 377-382 (2003). (article)
6. Fernández, C., Hilty, C., Wider, G. and Wüthrich, K. Lipid-protein interactions in DHPC micelles containing the integral membrane protein OmpX investigated by NMR spectroscopy. P. Natl. Acad. Sci. USA 99(21): 13533-13537 (2002). (article, web: Oct 7, 2002)
5. Hilty, C., Fernández, C., Wider, G. and Wüthrich, K. Side chain NMR assignments in the membrane protein OmpX reconstituted in DHPC micelles. J. Biomol. NMR 23(4): 289-301 (2002). (article)
4. Etezady-Esfarjani, T., Hilty, C., Wüthrich, K., Rueping, M., Schreiber, J. and Seebach, D. NMR-Structural Investigations of a β3-Dodecapeptide with Proteinogenic Side Chains in Methanol and in Aqueous Solutions. Helv. Chim. Acta 85(5): 1197-1209 (2002). (article, web: May 29, 2002)
3. Winterhalter, M., Hilty, C., Bezrukov, S.M., Nardin, C., Meier, W. and Fournier, D. Controlling membrane permeability with bacterial porins: application to encapsulated enzymes. Talanta 55(5): 965-971 (2001). (article, web: Nov 27, 2001)
2. Fernández, C., Hilty, C., Bonjour, S., Adeishvili, K., Pervushin, K. and Wüthrich, K. Solution NMR studies of the integral membrane proteins OmpX and OmpA from Escherichia coli. FEBS Letters 504(3): 173-178 (2001). (article, web: Aug 28, 2001)
1. Hilty, C. and Winterhalter, M. Facilitated substrate transport through membrane proteins. Phys. Rev. Lett. 86(24): 5624-5627 (2001). (article, Jun 11, 2001)